SNPs: Human scgf
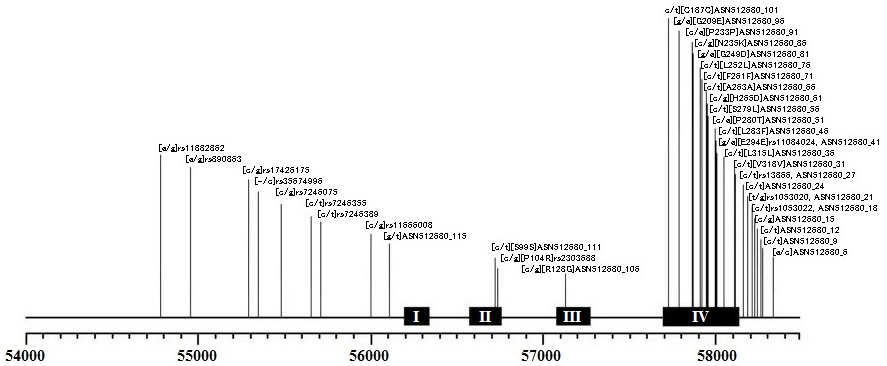
Total 35 SNPs ofscgf gene are deposited in Entrez SNP at NCBIand dbProP; 9 SNPs in the 5' flanking region, 2 SNPs with
1 synonymous and 1 nonsynonymous aa change in the 2nd exon, 1 SNP with nonsynonymous aa change in the 3rd exon, 15 SNPs
with 8 synonymous and 7 nonsynonymous aa changes in the 4th exon and 8 SNPs in the 3'-UTR. Locus and nucleotide change of
each SNP is shown below, where accession number follows [original/replaced nucleotide] with/without [A#A for synonymous aa
change or A#B for nonsynonymous aa change] in the coding region.
Frequent SNPs in the 4th exon and 3'-UTR actually do not misconduct SCGF molecule, implicating that the relevant area is
less associated with its biologic activity.
There have been some reports concerning correlation of
scgf SNPs with disease;
(1) no correlation of
scgf SNPs with cytogenetic response in chronic myeloid leukemia patients treated with imatinib (
18),
(2) significant correlation of schizophrenia with
scgf SNP
[a/g]rs722036 in the 3'-UTR (shown in the genome sequence), which
position further 1,566 nucleotides downstream fro
m [a/c]ASN512680_6 (Pat4).
(3) SNP (
[c/t]rs13866) of
scgf gene is associated with susceptibility to vitiligo vulgaris (
203). Vitiligo vulgaris is a depigmentary
disease caused by extinction of melanocytes. Keratinocytes play a homeostatic role for epidermal-melanin unit by elaborating KL,
bFGF, endothein-1, HGF and SCGF. Gene abnormality in the keratinocyte-derived growth factors has been one of the
pathogenetic candidates of vitiligo vulgaris. The keratinocyte-derived growth factor genes from 51 cases of vitiligo vulgaris without
autoimmune disease were compared with those from 118 normal controls.
KL SNP (rs11104947: G/A) and
scgf SNP (
[c/t]rs13866)
were associated with a 1.95- and a 2.14-fold higher susceptibility to vitiligo vulgaris, respectively, and the combined two gene SNPs
further showed a 7.92-fold higher risk of developing the disease.
Scgf -independently or
KL-dependently regulates a keratinocyte-melanocyte proliferative interaction, and
scgf-dysregulation is associated with susceptibility to vitiligo vulgaris, for (1)
scgf gene is expressed in neonatal or immature keratinocytes, (2)
scgf SNP (
[c/t]rs13866) is located at 3'UTR that is the target locus for hsa-miR-663, important for translation of
scgf mRNA, and (3)
scgf chromosomal locus 19q13.3 has been reportedly associated with vitiligo vulgaris. Association of vitiligo with
scgf SNP
[c/t]rs7246355 is in conflict; positive (
203) and negative (
303).
(4) Suppression of SCGF production by malarial pigment hemozoin-phagocytized monocytes is one of the causes underlying childhood severe malarial anemia (SMA<Hb 6.0g/dl) (
175). To further clarify a functional role of SCGF in SMA, it was investigated whether
scgf SNP
[c/t]rs7246355 was associated with the development of SMA in children with
P. falciparum malaria.
Scgf
genotyping indicated a significant prevalence of TT allele in non-SMA (6.0g/dl≤Hb<11.0g/dl) and CC allele in SMA. SCGF levels in the plasma and cultural supernatants of PB-MNCs from
P. falciparum-infected children with TT allele were higher than those with CC allele. In addition, TT allele groups had high reticulocyte production index relative to CC allele groups. Consequently homozygous T allele in the
scgf promoter (SNP
[c/t]rs7246355) at the position of 539 nucleotides upstream from the first exon exhibits resistance to the development of childhood severe malarial anemia (
221).
(5) A synonymous SNP [c/t] in the 2nd exon of
scgf gene is found in the tumor tissue from a patient with Her2
++, ER
-, PR
-ductal breast carcinoma (
377).
(6) Bioinformatics with ingenuity pathway analysis disclose that
scgf is one of the Erk1/2-linked gene SNPs commonly shared in alcohol, smoking and opioid addiction (
503).
(7)
F5 (coagulation factor V)
rs4656185,
STAB2 (stabilin 2)
rs187503377 and
scgf rs116924815 are associated with higher
circulating SCGF levels (
542).
Scgf Genome
54001 acaaggcctt tccccacata cgcattattt cttgaggtct gtagctcctg gtgaggggcc
54061 ccaggttccc gatgggcacc tgagaggccc aggctctgag gcagggagca gtctgcccaa
54121 ggtcacagtg ccctcgatgt accaggattt cttggaaaac ggaaggctgg aggcactgat
54181 ttccaaggcc cctcctgggg ctcccggcag ccgagggcag atgccctgct cttggagcca
54241 gaaaccttgg ctagctgccg ccacactggt gacctccagc aggctatgct gtttcagagc
54301 ctcggtctcc tcctttgaag agcaggccca gtgatccttg gctggtgtgc cccatgcttg
54361 ttggcaccac cagccaggcc tgggcgggaa aagccctgca tgactcatgg gcctgacata
54421 gaggggcatg gcatggcatt gttcttcctt gcctccaagc ctctctccca ctgccctcac
54481 cacctctacc taccctgcca ccaccaccct gggaacttgg gtgccagcaa accaaagagg
54541 tgaagcttcc aggcctttga ccccaccagg aacggctacc ctctcgccca ctcaacaggt
54601 tgattgaaca cctactatgt gctaggtagg ccttgtgcta ggagatgcat cagtgcataa
54661 aaatccctgc tttcattggc agggaacaga cataaacaaa caagatcatg ccagtcttga
[a/g]rs11882862
54721 ttataccaaa aagatgaaat ggggcatttt aataaagtaa cttgcggaat agataaggtg
54781 gttggggaag acctcctgga gctgagacct cgatgatgag aaggagctgg ggaaagaggc
54841 ttctagaagg aggaaagggc tagtgcaaat gctctgaggt tgtaatcagc ttagtatttg
[a/g]rs890863
54901 gggatgagcc aagaggccag ggaaggtgga ggctgggggt gggtgaaggg aatgggggtg
54961 atgaggctgc agaggtgcaa agggttcttc cttcccagcc cccagcatgg taggctgcgc
55021 tgaggactga ggcccgggct tggcttccct cctgcttatg atgggaaggt gggggagcag
55081 gggagtgggg agtgatgtga tctgatttat ggtttggaag gattgctttg gctgcggcaa
55141 gcagccggga ttggaggggc tcaggaggaa ggagggggag ggcgagactg cagtggtcca
55201 ggctgaaatg aggtggcttg ctgaatgctc agagctcctt gagcacacaa cctagcgtct
[c/g]rs17426175
55261 gtcgatttca cctcctggag aaaccttcccxtgactgctcc gccagactcc tcaccctctc
[-/c]rs35674996
55321 ccatctccag ccccctcatc cccggagcta tctggctgct ggtctgcaag cagctggagt
55381 ccccagcatc acccagccca cagcctggcc tgcaggaggc ttctagtaaa gtggccaatt
[c/g]rs7246075
55441 aatccccgag ccccgtcttc cagggtcatg cacacagtcaxcacgcgtgct cttcacatgt
55501 gtcaccctgc ccccacacac actcacttgg atctccagac aagggcttgg agggtaggat
55561 gggcaatcta gggggcagga ggggccctgg agtctcctcc attgagtgtg caggcgatgg
[c/t]rs7246355
55621 tggccacagg gtaataacaa cctcagcgag accctgaggt ctgcccctcc ttaggaatgg
[c/t]rs7246389
55681 ccaggcaggg aagaagggag cagtgctcgg tcccagcatc ccaccactgc cgagatggtt
55741 ctgagcgggt tcctgggctg aggccggggc ctgtgttttc caagattcct tggagagcca
55801 gctccatcct cttcctgatt atctgatctt tgggctacag cagaaggaaa gagcagggcg
55861 gaagcgagct ctggaaagtg gaaagaggag ggtgtgcgag gagacccaag gcgggatcta
55921 ggctttggaa cgttggaggt ttccggccac agaggcaggg cctccgggga gccaagaggg
[c/g]rs11666008
55981 gcggggagag acacacacag agaagcagag gagaccaacg gaccggacag agacgaggag
56041 aggaacagga agagagaagc tgggagaatc gggaacctgg gggctagtga cctgcacaca
[g/t]ASN512680_115
56101 gggcaggggc actcggcagt tcccagaggc cacccctccc accccagaca tccagacatc
56161 tggaactttg ggtgccaaga gtccagcttaxatgcaggcag cctggctttt gggggctttg 1st exon
56221 gtggtccccc agctcttggg ctttggccat ggggctcggg gagcagagag ggagtgggag
56281 ggaggctggg gaggtgccca ggaggaggag cgggagaggg aggccctgat gctgaaggtg
56341 agctctggag tgtcagggag ctatgggtgg tgaactgtgg gtctgggagg gggtattgca
56401 aggttaggga aatggacaat gaggagcgat tgggatagtc tcagaggagt gggagggtga
56461 tgggctcaga gacaggactc aggcagcaga gaatgagtta tgagttggaa aggggtgaat
56521 ggggcatctt ggtggcaggc tggggctcac cacccctccc ctgccccagc atctgcagga 2nd exon
56581 agccctagga ctgcctgctg ggagggggga tgagaatcct gccggaactg ttgagggaaa
56641 agaggactgg gagatggagg aggaccaggg ggaggaagag gaggaggaag caacgccaac
[c/t][S99S]ASN512680_111 [c/g][P104R]rs2303688 [c/t](377)
56701 cccatcctcc ggccccagcc cctctcccac ccctgaggac atcgtcactt acatccgtga
56761 gtaaccagaa gccctcaccc ccaaactcct aggccgccgc ggtcactttc gcaaaaatga
56821 agggtcggtt cactgccaag tggcctttca gttgtccatt gcccagccca gagcccccac
56881 acccctagga gccccaggtc cacggccatg cctcctaaag gcctgggcca ctcgacccta
56941 ccttccagga gtccggggtg ccccccccac cgccaccccg ccctcaggag ccctagatcc
57001 cagttctcca ggtcctcagg ccccccgctg catctccagg ctccccctcc agcatgatcc
57061 ctgtctgtcc gcagtgggcc gcctggccgg cctggacgca ggcctgcacc agctgcacgt 3rd exon
[c/g][R128G]ASN512680_106
57121 ccgtctgcac gcgttggaca cccgcgtggt cgagctgacc caggggctgc ggcagctgcg
57181 gaacgcggca ggcgacaccc gcgatgccgt gcaagccctg caggaggcgc agggtcgcgc
57241 cgagcgcgag cacggccgct tggagggtga gtccgcggcg cgcggggtgg aaaaaaatga
57301 gactatctgg gccaggaatg ggactggttg agaaggtgac cctaggtgtc cggggcggga
57361 gagttcggga gagctgctgt gtgctagcgg acggggttag agagctggga ggctgaatgc
57421 agggagcggg tgggcactcc agacccgcgc ggacccgact ggggcagcac cacgccccct
57481 acagtccagc tcccaccgcc tggccccgcc cctggcggac cagactctcc acctcctggc
57541 tcccgcggtt ctgaccacgc ctcctcccag ccctccccat gagttcctgg ccccgcctcc
57601 ctccaccccc ggatgttttg tcctcgcccc cttccagact ctgaatgcat gaccccgcct
57661 ccttctctac ccggccccgc ccacaggctg cctgaagggg ctgcgcctgg gccacaagtg 4th exon
[c/t][C187C]ASN512680_101
57721 cttcctgctc tcgcgcgact tcgaagctca ggcggcggcg caggcgcggt gcacggcgcg
[g/a][G209E]ASN512680_96
57781 gggcgggagc ctggcgcagc cggcagaccg ccagcagatg gaggcgctca ctcggtacct
[c/a][P233P]ASN512680_91[c/g][N235K]ASN512680_86
57841 gcgcgcggcg ctcgctccct acaactggcc cgtgtggctg ggcgtgcacg atcggcgcgc
[g/a][G249D]ASN512680_81[c/t][L252L]ASN512680_76[c/t][F261F]ASN512680_71[c/t][A263A]ASN512680_66[c/g][H265D]ASN512680_61
57901 cgagggcctc tacctcttcg aaaacggcca gcgcgtgtcc ttcttcgcct ggcatcgctc
[c/t][S279L]ASN512680_56[c/a][P280T]ASN512680_51[c/t][L283F]ASN512680_46
57961 accccgcccc gagctcggcg cccagcccag cgcctcgccg catccgctca gcccggacca
[g/a][E294E]rs11084024,ASN512680_41
58021 gcccaacggt ggcacgctcg agaactgcgt ggcgcaggcc tctgacgacg gctcctggtg
[c/t][L315L]ASN512680_36[c/t][V318V]ASN512680_31
58081 ggaccacgac tgccagcggc gtctctacta cgtctgcgag ttccccttct agcggggccg
[c/t]rs13866,ASN512680_27 [c/t]ASN512680_24
58141 gtaccccgcc tccctgccca tcccaccacc cggcctttccxctgcgccgtg cccaccctcc
[t/g]rs1053020,ASN512680_21[c/t]rs1053022,ASN512680_18 [c/g]ASN512680_15 [c/t]ASN512680_12
58201 tccggaatctxcccttcccttxcctggccacg aatggcagcg tcctccccga cccccagtct
[c/t]ASN512680_9
58261 gggcgcttct gggagggctc ttgcggtgcc ggcactcctc cttgttagtg tctttccttg
[a/c]ASN512680_6
58321 aaggggcggg caccaggcta ggtccggtgc caataaatcc ttgtggaatc tgacttgagg
58381 ggcagtgaag gcagtcttgg actttatctc ctgcagggga agccgggaag ggccagaccc
58441 tggtctttgg gcgcgtgtcc tggcctttgg atgcctgttc tgctttttac agatgtcatt
58501 tgagccccag atggagtgat ggagaaacac aggtgcatat gtacacacgg tcatctgttc
58561 tgtgcacaga tggccggagt acacgcgtac aaacgctgta gacaccttca cctccatcgt
58621 gccctctctc tcggcctttg ggacccaggg cctgccagtt gtcccaggcc ttggtcttgg
58681 tcctggtcaa agagatttgg gaaacgtgaa ggggctaaag gctctgacaa gtccggcagg
58741 aaagagcccg tttaatttgt ttaagtggcg tgtccaattt gtttccccag atgatatggt
58801 ttcatggact tgttgggatg atatattgag acatggactt aaggactcac ccaaccctga
58861 agcttagact tcctcccaat tttccctcct ctgcttcctg ccagtgaggc caggagccag
58921 tcccttcccc tctgtgagcc tcgggttttt catctgaacg tgggcacaat ccctcctact
58981 ttgtgcggct cttgggatgt tcaggtgaga ccaggacagt gtgcatttgg tggtggctga
59041 acttgagcca cacactacct tccgtcctca cctttttttt tccttttttg agacacggtc
59101 ttgctctttc gcccaggctg gagtgcagtg gtacaatcat ggcttactgc agcctcaatc
59161 ttctgggttc aagcagtcct cccaccttag ccccccaagt agctgggact acaggcacgc
59221 cccaccacac ctggctaatt tttgtatttt tttatagagg ttggttttcg ccacgttgct
59281 caggctggtc ttgaactcct gagctcaagc aatctgccct cctgggcctc ccaaagggct
59341 gggaatacag gcatgagcca cagtgcctgg ctctccctca ccatttttca tacctttact
59401 ccctgggggc agtgaagaca cccaggagtc cgccagccgg ggcctcagca tagtggccag
59461 ctctgtctag aggagaggct ctttggaaat caccctagta cctcccccaa ggttggcatc
59521 atgggcgtga ccctgacatc agggagccta ggaatgggga tgtgaccctg tctgtcctta
59581 ccactctgcc ccttcccttc cttcagccat tgcaccactg cctaaagggt gaccattgga
59641 tccacacatg ggaggagcaa gccatcactg tcagcttgga tagaggggag atcacccacc
59701 cttctccctt caggagacat ttctcagccc cacatcccca gaggaataca gaataggaga
59761 gtgggggtgt ggcctggagt cagccttcca tcagaaattc ttctcatgct gagcttcagt
59821 ttccccgggg agggataggc agccctagat cttatagcca accagaggtc agattaggca
[a/g]rs722036
59881 gacaatgatg agtatctatt atttcccaga ggggtctctg ccctggcaga gcttccattc
59941 tggaaagtat ttgggcatgg tacaccatgg ctacccgccc agattctaga gccagatggc
SNPs: Mouse scgf
Mouse interstrain SNPs are listed at
Mouse Genome Informatics (MGI).